| x1 | x2 | x3 | x4 | x5 | x6 | x7 |
|---|---|---|---|---|---|---|
| 0.3981105 | 13.912435 | a | 0 | -0.6775811 | 0.8759740 | -0.2051604 |
| -0.1434733 | 1.093743 | c | 0 | 0.7055193 | 0.2521987 | 1.8816947 |
| -0.2526000 | 4.898035 | c | 0 | 0.4744651 | -0.5628840 | 0.3245589 |
| -1.2272588 | 14.717053 | b | 0 | -0.5132792 | -1.1368242 | -0.1355150 |
| -0.4360417 | 8.547025 | c | 1 | -0.1736804 | -0.7120962 | -1.2714320 |
Reproducible Science
Replicability School
June 6, 2025
About me 👋
- Post-doctoral researcher in Cognitive Psychology, University of Padova.
- Research: Computational modeling of cognitive and learning processes, Bayesian hypothesis testing.
- PhD in Psychological Science, completed March 6, 2025.
- Passionate about reproducible science after struggling with disorganized datasets in my early research!
Our job is hard 🔥
Running experiments
Analyzing data
Managing trainees
Writing papers
Responding to reviewers

Reproducibility helps!

Organizes your workflow.
Saves time by documenting steps.
Builds trust in your findings.
Enables others to reproduce and extend your work.
What is reproducible science?
At its core, reproducible science means that someone else, or even you, in the future, can reproduce your results from your materials: your data, your code, your documentation.
It means your workflow is transparent.
Keys to reproducible science 🔐
- Data: organize, document, and share your datasets in ways that are usable by others and understandable by you (even years later).
- Code: write analysis scripts that are clean, transparent, and reusable..
- Literate programming: combine code and text in the same document, so your reports are dynamic and replicable.
- Version Control and Sharing: track changes, collaborate, and make your work openly available using tools like GitHub and OSF.
So… Is reproducible science even harder?
At first, yes - but then…🧯🔥
- Helps you stay organized.
- Makes it easier to remember what you did.
- Allows others to understand, reproduce, and build on your work.
Learning the tools takes effort but once you do, your workflow becomes smoother, clearer, and more reliable.
Outline
Data
Code
R projects
Literate Programming
Version Control
Data
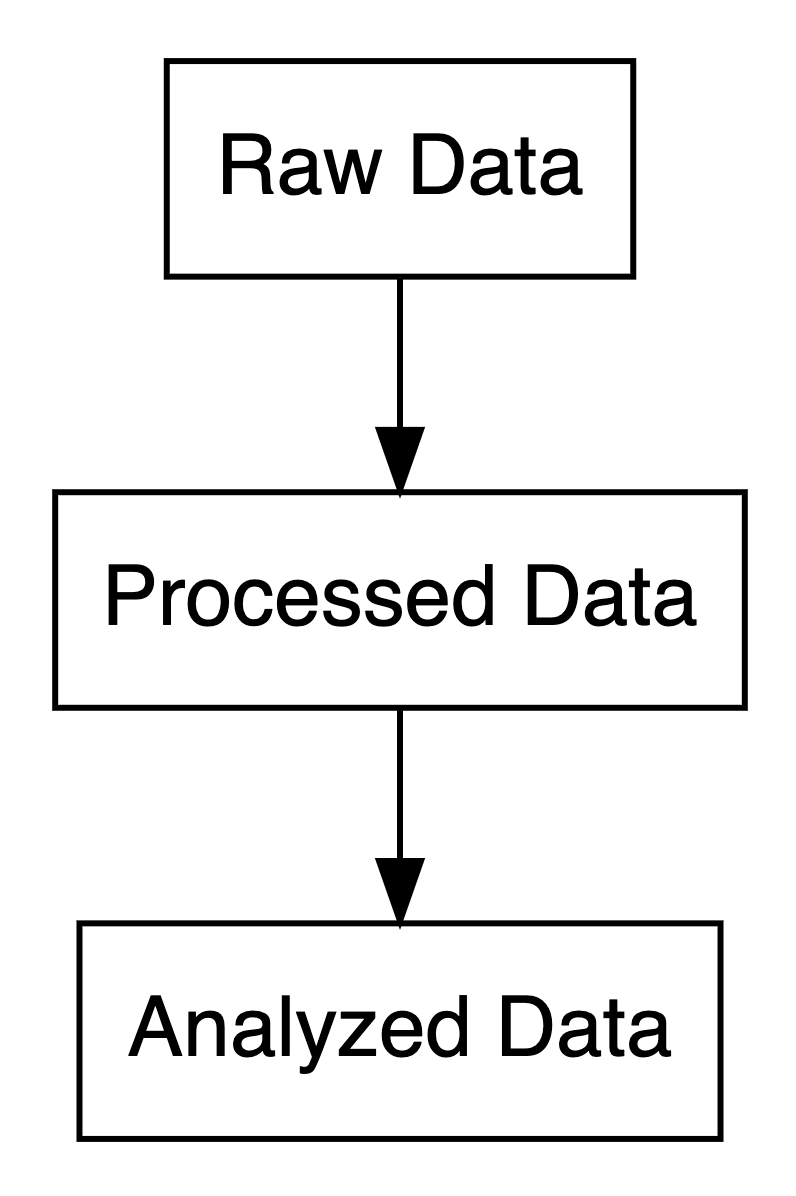
Data types in research
- Raw Data: Original, unprocessed (e.g., survey responses).
- Processed Data: Cleaned, digitized, or compressed.
- Analyzed Data: Summarized in tables, charts, or text.
Open Science Framework
- Free platform to organize, document, and share research.
- Supports preregistration, archiving, and collaboration.
- Integrates with GitHub, Dropbox, Google Drive.
Bad data sharing example
Imagine this scenario: you read a paper that seems really relevant to your research. At the end, you’re excited to see they’ve shared their data on OSF. You go to the repository, and there’s one file…
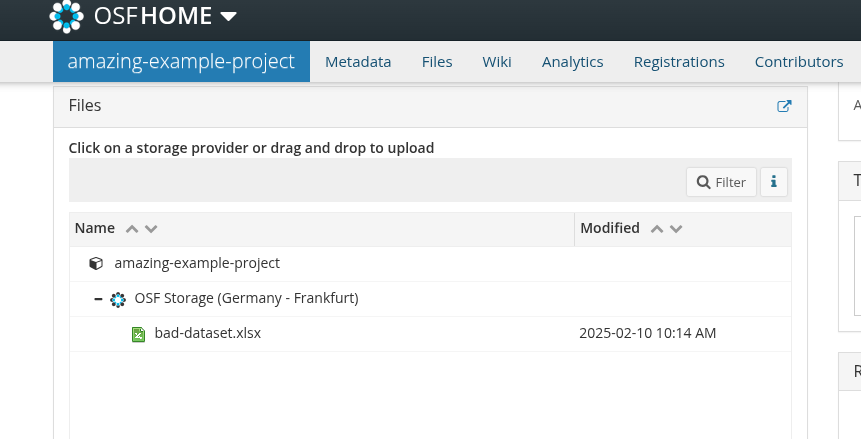
Bad data sharing example
You download it, open it, and you see this: . . .
What do these variables mean? What’s x3?
What do 0 and 1 represent? How are missing values coded?
Is x6 a z-score or raw data?
Good data sharing practices
- Use plain-text formats (e.g.,
.csv,.txt). - Include a data dictionary with variable descriptions.
- Add a README with key details.
- Follow FAIR principles (Findable, Accessible, Interoperable, Reusable).
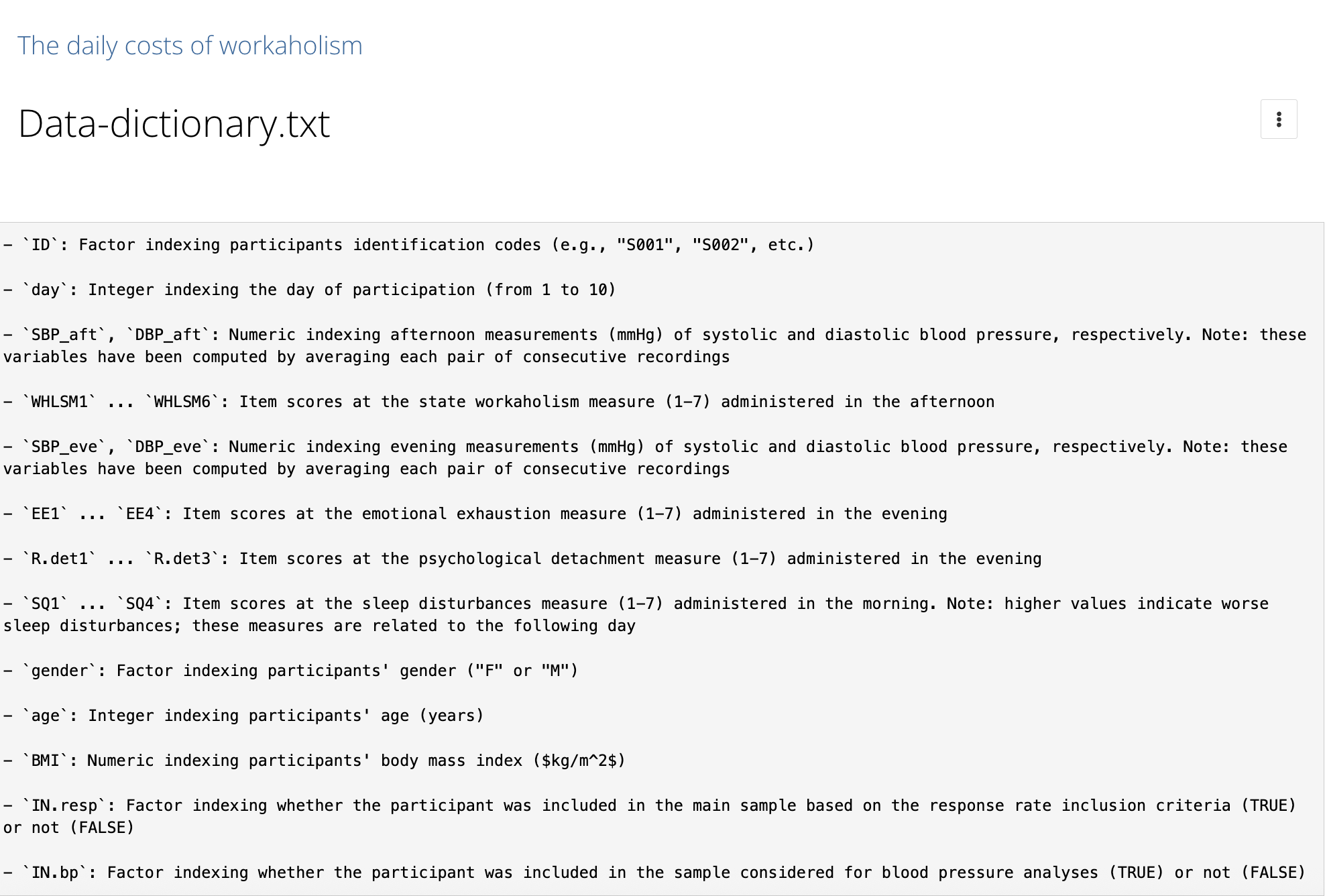
Data dictionary 📂
- A data dictionary defines each variable in your dataset.
- Boosts transparency and collaboration.
- Saves time for collaborators and future-you.
datadictionary 📦
library(datadictionary)
df <- data.frame( id = factor(letters[1:5]),
anxi = rnorm(5, 0, 1),
edu = factor(c("PhD", "BSc", "MSc", "PhD", "BSc")))
df_labels <- list(
anxi = "Beck Anxiety Inventory, standardized",
edu = "Last degree obtained"
)
create_dictionary(df, id_var = "id", var_labels = df_labels) item label class summary value
1 Rows in dataset 5
2 Columns in dataset 3
3 id Unique identifier unique values 5
4 missing 0
5 anxi Beck Anxiety Inventory, standardized numeric mean 0
6 median 0
7 min -1.03
8 max 0.22
9 missing 0
10 edu Last degree obtained factor BSc (1) 2
11 MSc (2) 1
12 PhD (3) 2
13 missing 0Data dictionary - Good data sharing example
README files
A README file is the first thing someone sees when they open your dataset/project folder. It should answer basic questions like:
- What is this dataset?
- How was it collected?
- What are the variables?
- Which is the structure of the project?
README - Good data sharing example
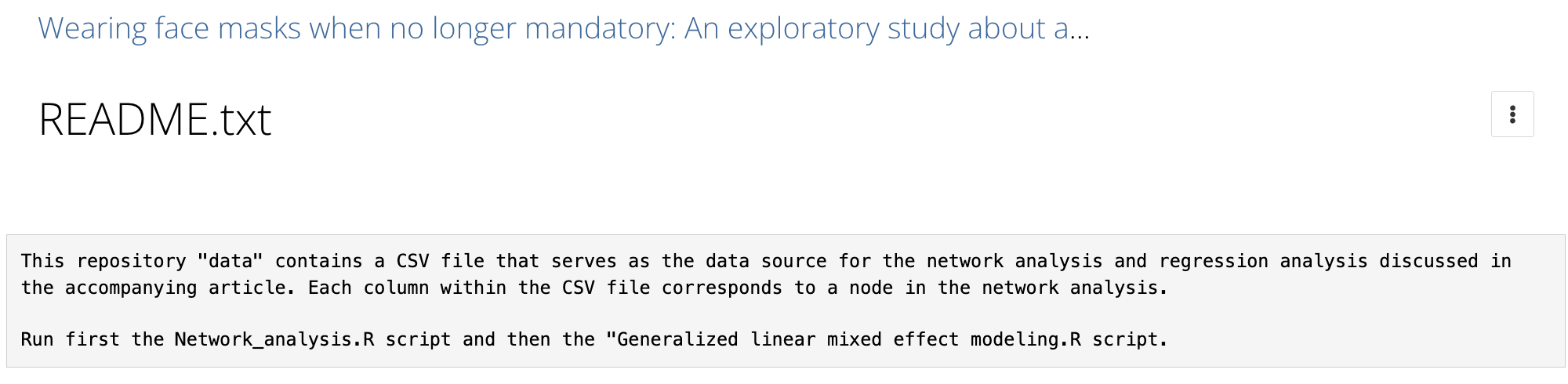
README - Good data sharing example

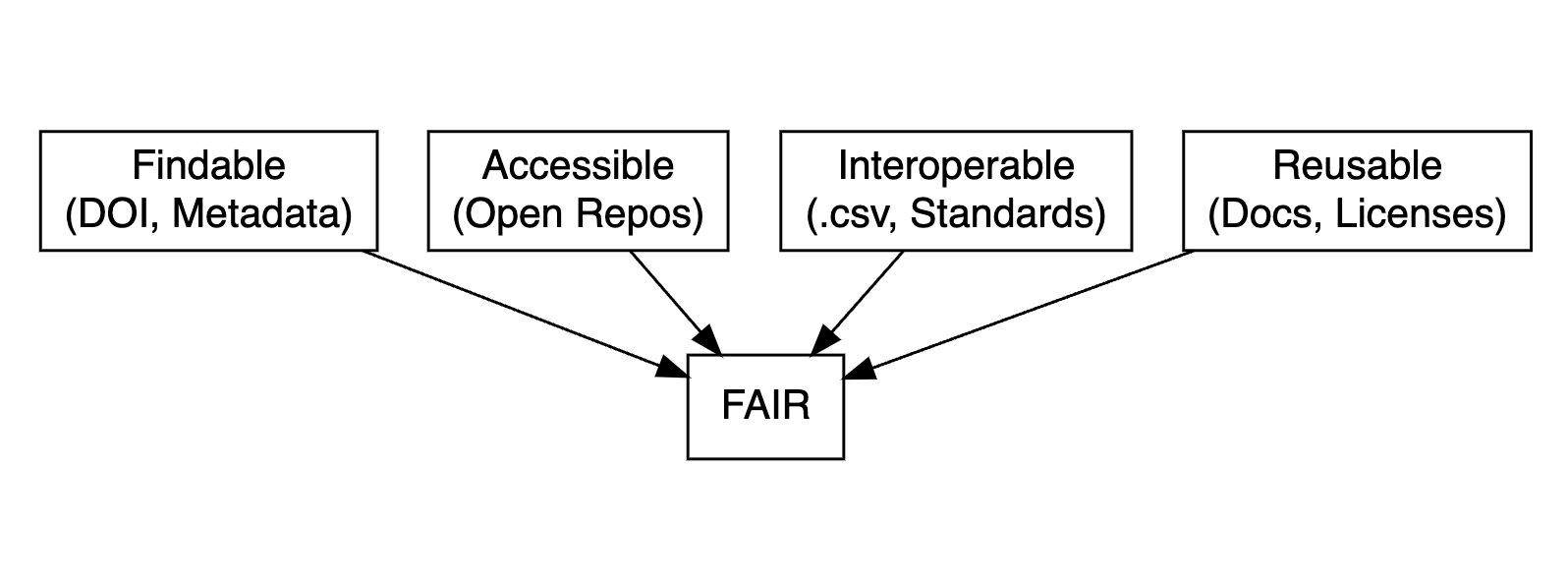
FAIR data principles 🔍
- Findable: Use metadata and DOIs to make data easy to locate.
- Accessible: Ensure data is retrievable via open repositories.
- Interoperable: Use standard formats (e.g.,
.csv,.txt) for compatibility. - Reusable: Include clear documentation and open licenses.
Data licensing 🔒
A license tells others what they can and can’t do with your data. If you don’t include one, legally speaking, people might not be allowed to use it, even if you meant to share it openly.
- Licenses clarify how others can use your data.
- Common licenses:
Code
Why scripting?
The SPSS Workflow
Click menu items to run analysis
“exclude <18”
Click through everything again
Forget a step? Round differently?
Stressful, error-prone, and undocumented.
Why scripting?
Scripting ensures transparent and reproducible workflows.
Reproducible: You can rerun them.
Documented: You can see what you did and when.
Shareable: Others can inspect and reproduce your analysis.
R and RStudio 💻
- R: Free, open-source, with thousands of packages for analysis.
- RStudio: Intuitive interface for coding, plotting, and debugging.
- Vibrant community for support and resources.
Writing better code 📝
- Name descriptively: Use
snake_caseorcamelCasefor readability. - Comment clearly: Document your logic for clarity.
- Organize scripts: Load packages and data upfront.
Use descriptive names
Another best practice: name your variables clearly.
Consistency helps too. Use either snake_case or camelCase, but pick one and stick to it.
Organized scripts
Global operations at the beginning of the script:
- loading packages
- loading datasets
- changing general options (
options())
Summary
- Scripts beat point-and-click
- Structure matters
- Comment often
- Name things well
Functions to avoid repetition
Functions are the primary building blocks of your program. You write small, reusable, self-contained functions that do one thing well, and then you combine them.
Avoid repeating the same operation multiple times in the script. The rule is, if you are doing the same operation more than two times, write a function.
A function can be re-used, tested and changed just one time affecting the whole project.
Functional Programming, example…
We have a dataset (mtcars) and we want to calculate the mean, median, standard deviation, minimum and maximum of each column and store the result in a table.
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1'data.frame': 32 obs. of 11 variables:
$ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
$ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
$ disp: num 160 160 108 258 360 ...
$ hp : num 110 110 93 110 175 105 245 62 95 123 ...
$ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
$ wt : num 2.62 2.88 2.32 3.21 3.44 ...
$ qsec: num 16.5 17 18.6 19.4 17 ...
$ vs : num 0 0 1 1 0 1 0 1 1 1 ...
$ am : num 1 1 1 0 0 0 0 0 0 0 ...
$ gear: num 4 4 4 3 3 3 3 4 4 4 ...
$ carb: num 4 4 1 1 2 1 4 2 2 4 ...Imperative Programming
The standard (~imperative) option is using a for loop, iterating through columns, calculate the values and store into another data structure.
ncols <- ncol(mtcars)
means <- medians <- mins <- maxs <- rep(0, ncols)
for(i in 1:ncols){
means[i] <- mean(mtcars[[i]])
medians[i] <- median(mtcars[[i]])
mins[i] <- min(mtcars[[i]])
maxs[i] <- max(mtcars[[i]])
}
results <- data.frame(means, medians, mins, maxs)
results$col <- names(mtcars)
head(results, n = 3) means medians mins maxs col
1 20.09062 19.2 10.4 33.9 mpg
2 6.18750 6.0 4.0 8.0 cyl
3 230.72188 196.3 71.1 472.0 dispFunctional Programming
The main idea is to decompose the problem writing a function and loop over the columns of the dataframe:
Functional Programming
Functional Programming, *apply 📦
- The
*applyfamily is one of the best tool in R. The idea is pretty simple: apply a function to each element of a list. - The powerful side is that in R everything can be considered as a list. A vector is a list of single elements, a dataframe is a list of columns etc.
- Internally, R is still using a
forloop but the verbose part (preallocation, choosing the iterator, indexing) is encapsulated into the*applyfunction.
The *apply Family
Apply your function…
Now results is a list of data frames, one per column.
We can stack them into one big data frame:
means medians mins maxs
mpg 20.090625 19.200 10.400 33.900
cyl 6.187500 6.000 4.000 8.000
disp 230.721875 196.300 71.100 472.000
hp 146.687500 123.000 52.000 335.000
drat 3.596563 3.695 2.760 4.930
wt 3.217250 3.325 1.513 5.424
qsec 17.848750 17.710 14.500 22.900
vs 0.437500 0.000 0.000 1.000
am 0.406250 0.000 0.000 1.000
gear 3.687500 4.000 3.000 5.000
carb 2.812500 2.000 1.000 8.000Using sapply, vapply, and apply
lapply()always returns a list.sapply()tries to simplify the result into a vector or matrix.vapply()is likesapply()but safer (you specify the return type).apply()is for applying functions over rows or columns of a matrix or data frame.
for loops are bad?
for loops are the core of each operation in R (and in every programming language). For complex operation thery are more readable and effective compared to *apply. In R we need extra care for writing efficent for loops.
Extremely slow, no preallocation:
Very fast:
microbenchmark 📦
library(microbenchmark)
microbenchmark(
grow_in_loop = {
res <- c()
for (i in 1:10000) {
res[i] <- i^2
}
},
preallocated = {
res <- rep(0, 10000)
for (i in 1:length(res)) {
res[i] <- i^2
}
}, times = 100)Unit: microseconds
expr min lq mean median uq max neval
grow_in_loop 1168.090 1234.7355 1446.4919 1276.2275 1393.3235 6538.393 100
preallocated 655.508 672.2975 717.7563 685.7045 715.2245 2393.990 100
cld
a
bGoing further: custom function lists
Let’s define a list of functions:
Now we can apply all of these to every column:
mean sd min max median
mpg 20.090625 6.0269481 10.400 33.900 19.200
cyl 6.187500 1.7859216 4.000 8.000 6.000
disp 230.721875 123.9386938 71.100 472.000 196.300
hp 146.687500 68.5628685 52.000 335.000 123.000
drat 3.596563 0.5346787 2.760 4.930 3.695
wt 3.217250 0.9784574 1.513 5.424 3.325
qsec 17.848750 1.7869432 14.500 22.900 17.710
vs 0.437500 0.5040161 0.000 1.000 0.000
am 0.406250 0.4989909 0.000 1.000 0.000
gear 3.687500 0.7378041 3.000 5.000 4.000
carb 2.812500 1.6152000 1.000 8.000 2.000This gives you a matrix with rows as variables and columns as statistics.
Pure vs. Impure functions
Test your functions - fuzzr 📦
When you write your own functions, it’s smart to test them. In R, we can use fuzzr to do property-based testing.
Define your function…
This runs the property on different random numeric vectors and checks whether it holds.
Why functional programming?
- We can write less and reusable code that can be shared and used in multiple projects.
- The scripts are more compact, easy to modify and less error prone (imagine that you want to improve the
summfunction, you only need to change it once instead of touching theforloop). - Functions can be easily and consistently documented (see roxygen documentation) improving the reproducibility and readability of your code.
Functional programming in the wild
You can write some R scripts only with functions and source() them into the global environment.
project/
├─ R/
│ ├─ utils.R
├─ analysis.RMore about functional programming in R
- Advanced R by Hadley Wickham, section on Functional Programming (https://adv-r.hadley.nz/fp.html)
- Hands-On Programming with R by Garrett Grolemund https://rstudio-education.github.io/hopr/
- Hadley Wickham: The Joy of Functional Programming (for Data Science)(https://www.youtube.com/watch?v=bzUmK0Y07ck)
Wrapping up
- Avoid repetition by using functions.
- Favor pure functions.
- Test your functions.
- The
*applyfunctions are your friends, or*mapfrompurrr📦
Organize your project
R Projects
R Projects are a feature implemented in RStudio to organize a working directory.
- They automatically set the working directory
- They allow the use of relative paths instead of absolute paths
- They provide quick access to a specific project
The Working Directory Problem
How many times have you opened an R script and seen this at the top?
setwd(“C:/Users/margherita/Documents/PhD/final_data/mess”) #change
Instead of hardcoding paths, we want to use projects with relative paths.
R Projects
An R Project (.Rproj) is a file that defines a self-contained workspace.
When you open an R Project, your working directory is automatically set to the project root, no need to use setwd() ever again.
Open RStudio
File → New Project → New Directory → New Project
Relative Path (to the working directory)
Users
|
├─tita
|
├─ workin_memo
|
├─ data
| ├─ clean_data.csv
|
├─ workin_memo.RprojAbsolute path: read.csv(“Users/tita/workin_memo/data/clean_data.csv”)
Relative path: read.csv(“data/clean_data.csv”)
A Minimal Project Structure
my-project/
│
├── data/
│ ├── raw/
│ └── processed/
├── R/
│ └── analysis.R
│
├── outputs/
│ ├── figures/
│ └── tables/
│
├── my-project.Rproj
│
└── README.mdProject organization with rrtools 📦
To make this even “easier”, you can use the rrtools package to create what’s called a reproducible research compendium.
… the goal is to provide a standard and easily recognisable way for organising the digital materials of a project to enable others to inspect, reproduce, and extend the research… (Marwick et al., 2018)
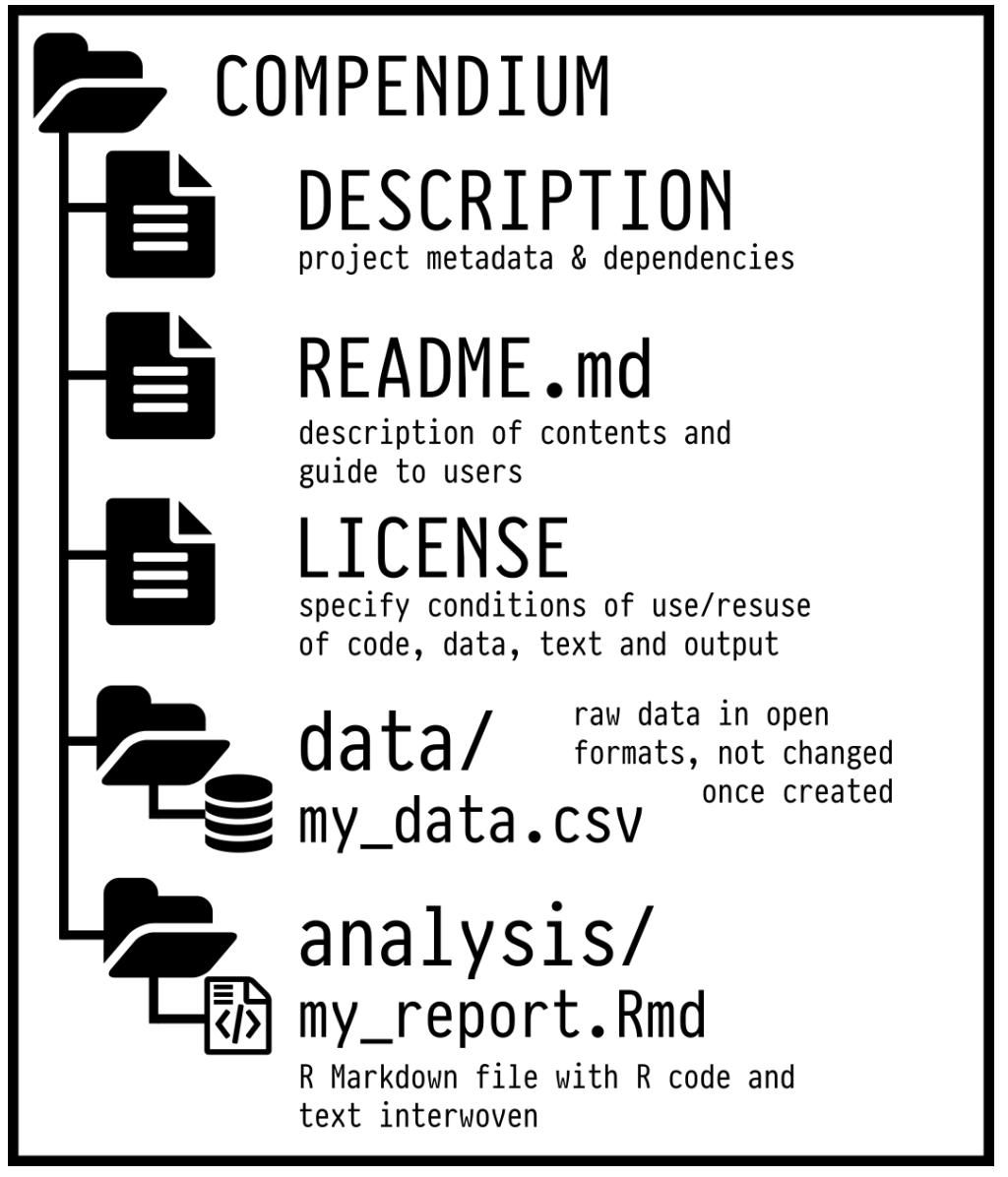
Research compendium rrtools 📦
- Organize its files according to the prevailing conventions.
- Maintain a clear separation of data (original data is untouched!), method, and output.
- Specify the computational environment that was used for the original analysis
rrtools::create_compendium("compedium") builds the basic structure for a research compendium.
renv📦 : locking your R environment
Another challenge for reproducibility is package versions.
You write some code today using dplyr 1.1.2.
In six months, dplyr gets updated… 😢
renv helps you create reproducible environments for your R projects!
What does renv do?
It records all the packages you use, with versions, in a lockfile
It installs them in a project-specific library
It ensures that anyone who runs your code gets exactly the same environment
Project specific library
install.packages("renv")
renv::init()
install.packages('bayesplot')
These packages will be installed into "~/repro-pre-school/example-renv/renv/library/macos/R-4.4/aarch64-apple-darwin20".renv commands
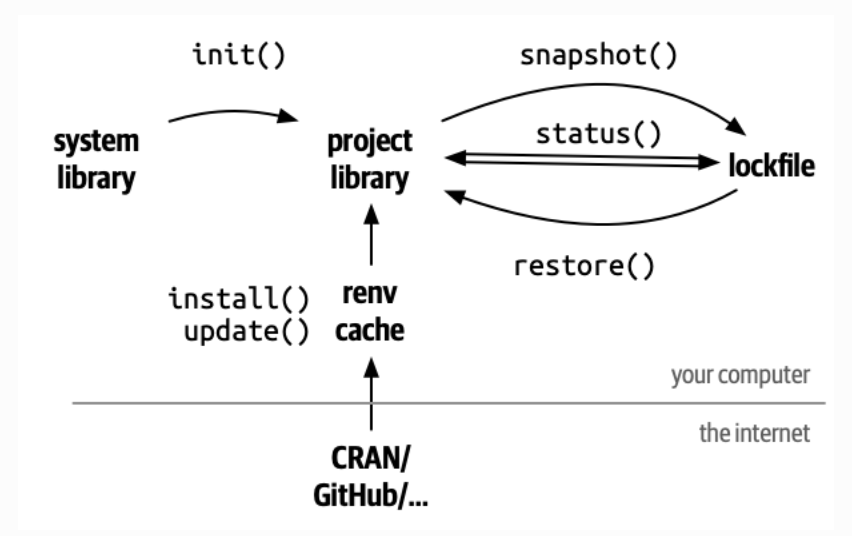
renv::restore() # re-install from lockfile
Research rrtools + renv 💣
rrtools: Organizes your project into a reproducible compendium with clear folders.renv: Locks R package versions for consistent environments.- Together, they ensure structure and reproducibility across teams and time.
- Run
rrtools::create_compendium("project")to start, thenrenv::init()to lock dependencies.
![]() Docker
Docker
- Packages your project’s software, dependencies, and system settings into a container.
- Ensures consistency across different computers or servers.
- Ideal for sharing complex analyses with others.
Documenting your environment ℹ️
sessionInfo(): Captures your R version, packages, and platform in one command.- Easy way to document and share your environment.
R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Sequoia 15.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Rome
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] fuzzr_0.2.2 microbenchmark_1.5.0 datadictionary_1.0.1
loaded via a namespace (and not attached):
[1] gt_1.0.0 sandwich_3.1-1 sass_0.4.10 generics_0.1.4
[5] tidyr_1.3.1 xml2_1.3.8 stringi_1.8.7 lattice_0.22-6
[9] hms_1.1.3 digest_0.6.37 magrittr_2.0.3 evaluate_1.0.3
[13] grid_4.4.2 timechange_0.3.0 mvtnorm_1.3-3 fastmap_1.2.0
[17] cellranger_1.1.0 jsonlite_2.0.0 Matrix_1.7-2 zip_2.3.3
[21] survival_3.8-3 multcomp_1.4-28 purrr_1.0.4 TH.data_1.1-3
[25] codetools_0.2-20 cli_3.6.5 labelled_2.14.1 rlang_1.1.6
[29] splines_4.4.2 withr_3.0.2 yaml_2.3.10 tools_4.4.2
[33] dplyr_1.1.4 forcats_1.0.0 assertthat_0.2.1 vctrs_0.6.5
[37] R6_2.6.1 zoo_1.8-12 lifecycle_1.0.4 lubridate_1.9.4
[41] MASS_7.3-64 pkgconfig_2.0.3 pillar_1.10.2 openxlsx_4.2.8
[45] glue_1.8.0 Rcpp_1.0.14 haven_2.5.4 xfun_0.52
[49] tibble_3.2.1 tidyselect_1.2.1 rstudioapi_0.17.1 knitr_1.50.4
[53] htmltools_0.5.8.1 rmarkdown_2.29 compiler_4.4.2 chron_2.3-62
[57] readxl_1.4.3 Organizing for reproducibility
- Don’t hardcode paths, use
R Projects - Create a logical folder structure for your project
- Use
rrtoolsto scaffold a research compendium - Use
renvto lock your package versions
Literate Programming
What’s wrong about Microsoft Word?
In MS Word (or similar) we need to produce everything outside and then manually put figures and tables.
- writing math formulas
- reporting statistics in the text
- producing tables
- producing plots

Think about the typical MW workflow
- You run your analysis in R
- You copy the results into a Word document
- You tweak the formatting
- You insert a figure generated with R manually
- You change your analysis, but forget to update the results in the text…
Literate Programming
A document where:
- The code is part of the text
- The results are generated dynamically
- The figures are rendered automatically
- Everything is in sync
For example jupyter notebooks, R Markdown and now Quarto are literate programming frameworks to integrate code and text.
Literate Programming, the markup language
The markup language is the core element of a literate programming framework. When you write in a markup language, you’re writing plain text while also giving instructions for how to generate the final result.
- LaTeX
- HTML
- Markdown
- …
LaTeX

Markdown
Markdown
Markdown is one of the most popular markup languages for several reasons:
- easy to write and read compared to Latex and HTML
- easy to convert from Markdown to basically every other format using
pandoc
Quarto
Quarto (https://quarto.org/) is the evolution of R Markdown that integrate a programming language with the Markdown markup language. It is very simple but quite powerful.
Basic Markdown
Markdown can be learned in minutes. You can go to the following link https://quarto.org/docs/authoring/markdown-basics.html and try to understand the syntax.
Quarto
You write your documents in Markdown, and Quarto turns them into:
- HTML reports
- PDF articles
- Word documents
- Slides
- Website
- Academic manuscripts
- …
Quarto
- If your data changes, your summary table updates.
- If you update your model, your coefficients update.
- If you change a plot’s colors, the new version appear, without having to re-export and re-insert anything.
This eliminates a huge source of human error: manual updates.
Outputs
Quarto can generate multiple output formats from the same source file.
- A PDF to send to your colleagues
- A Word document for your co-author who hates PDFs
- An HTML report for your own website
Everything from the same source. No duplication. Synchronization.
Extra Tools: citations and cross-referencing
- Citations with BibTeX or Zotero
- Cross-references for figures and tables
- Numbered equations with LaTeX syntax
- Footnotes, tables of contents, and more
You can write scientific documents that look and behave just like journal articles, without ever opening Word.
Writing Papers - APA quarto
APA Quarto is a Quarto extension that makes it easy to write documents in APA 7th edition style, with automatic formatting for title pages, headings, citations, references, tables, and figures.
Let’s see an example…
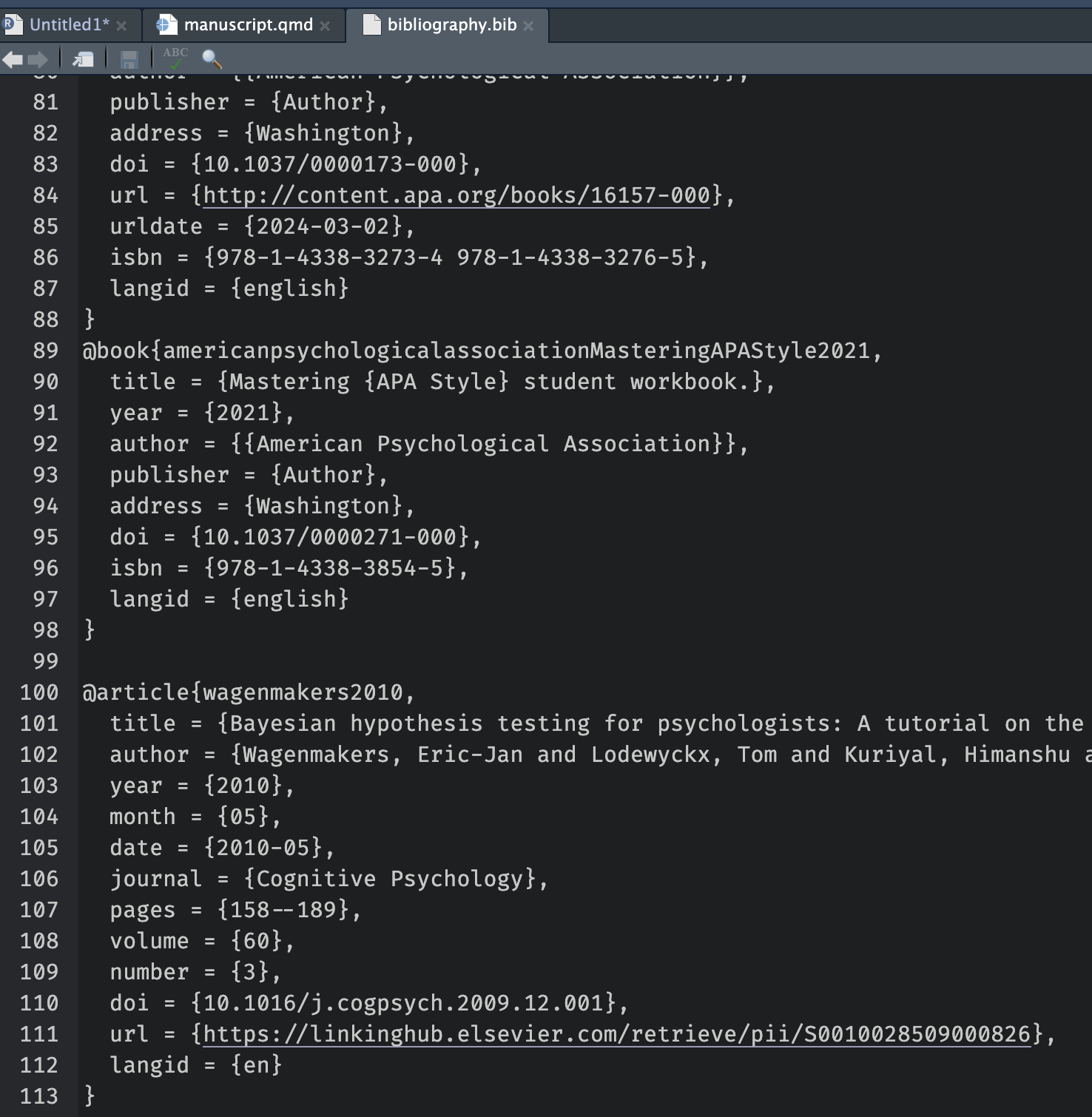
Quarto + Zotero ![]()
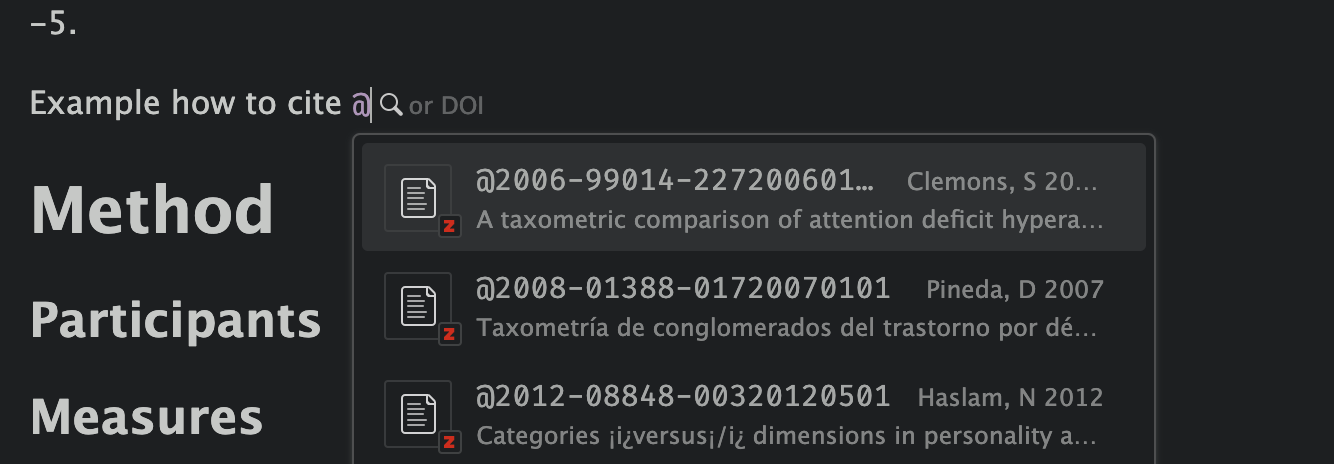
Choose your reference: 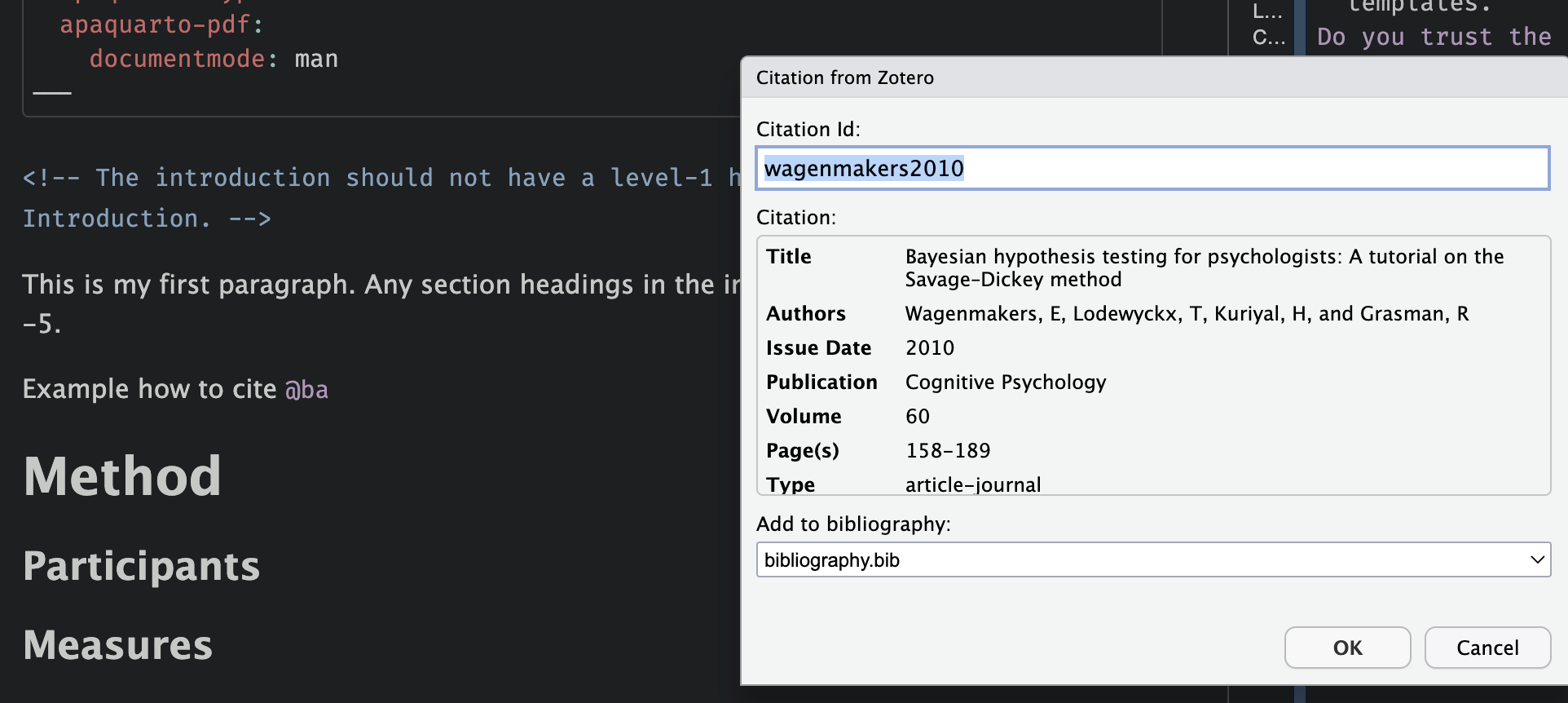
More about Quarto and R Markdown
The topic is extremely vast. You can do everything in Quarto, a website, thesis, your CV, etc.
- Yihui Xie - R Markdown Cookbook https://bookdown.org/yihui/rmarkdown-cookbook/
- Yihui Xie - R Markdown: The Definitive Guide https://bookdown.org/yihui/rmarkdown/
- Quarto documentation https://quarto.org/docs/guide/
Version Control
Why Version Control?
You’re working on a project. You save your script as:
analysis.Ranalysis2.Ranalysis_final.Ranalysis_final_revised.Ranalysis_final_revised_OK_for_real.R
What is Git?
Git is a version control system. It works like a time machine for your project.
Then, save changes with commits:
Commit message ✍️
- Write meaningful messages:
- ✅
"Fix bug in anxiety scoring function" - ❌
"stuff" - Use the imperative mood:
"Add README","Update plots"
GitHub
Git works locally. GitHub is the online platform for:
- Backing up your project
- Sharing it publicly or privately
- Collaborating with others
- Tracking issues and progress
GitHub in Practice
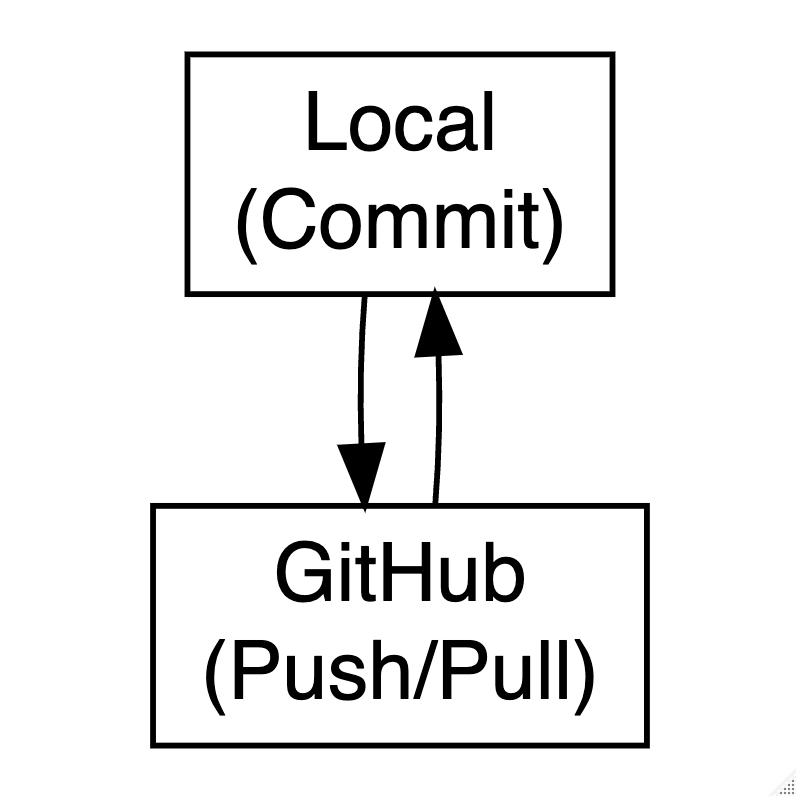
# 1. Initialize a Git repository in your current project folder
git init
# 2. Stage a file to be tracked (e.g., your script)
git add analysis.R
# 3. Save a snapshot of your work with a message
git commit -m "Initial commit"
# 4. Connect your local project to a GitHub repo (change the URL)
git remote add origin https://github.com/yourname/repo.git
# 5. Upload your commits to GitHub
git push -u origin mainBranching & merging 🌱
Try out new features
Fix bugs safely
Work on different versions in parallel
# Create and switch to a new branch called 'new-feature'
git checkout -b new-feature
# (Make your changes in code, then stage and commit them)
# Save those changes with a descriptive message
git commit -m "Add new plot"
# Switch back to the main branch
git checkout main
# Merge the changes from 'new-feature' into 'main'
git merge new-featureUse branches to keep your
mainbranch clean.
Handling conflicts
Sometimes, Git can’t automatically merge changes. This happens when two branches modify the same line in a file.
Git will insert conflict markers directly into the file:
The code between <<<<<<< HEAD and ======= is from the current branch (e.g., main)
The code between ======= and >>>>>>> new-feature is from the other branch you’re merging (e.g., new-feature)
Handling conflicts
To resolve the conflict, choose the correct version (or combine them), delete the markers, and save the file.
For example:
Then:
GitHub + RStudio Integration
- Clone repos with
File → New Project → Version Control - Commit and push from the Git tab in RStudio
- View commit history in History pane
Practice & resources
- Happy Git and GitHub for the useR
- GitHub Education: https://education.github.com
- Try GitHub Desktop (GUI client)
Start small. Use Git for one script. Then grow your skills from there.
If Git and GitHub feel too technical, or if your collaborators are less technical, the OSF is a fantastic alternative or complement.
- Upload data, code, and documents
- Create public or private projects
- Add collaborators
- Create preregistrations
- Generate DOIs for citation
- Track changes
You can also connect OSF to GitHub.
Integrated workflow 🛠️
- Develop your analysis using R and Quarto.
- Track code and scripts using Git.
- Host your code on GitHub (public or private).
- Upload your data and materials to OSF, including a data dictionary.
- Link your GitHub repository to your OSF project.
- Use
renvfor reproducible R environments.
- Share the OSF project and cite it in your paper.

Reproducibility
It’s about credibility and transparency.
Reproducible science is not about being perfect.
It’s about showing your work so that others can follow, understand, and build upon it.
Start simple, don’t wait until you’re “ready”, and teach what you learn!
THANK YOU!




Comments, comments and comments…
Write the code for your future self and for others, not for yourself right now.
Try to open a (not well documented) old coding project after a couple of years and you will understand :)
Invest time in writing more comprehensible and documented code for you and others.